See page 285 of textbook
- Plasmid - Small circular pieces of DNA that can exit and enter bacterial cells
- Plasmid Mapping - Process to determine where certain recognition sites are located and the number of recognition sites within the plasmid
- Copy Number - Number of copies of a particular plasmid found in a bacteria cell
- Multiple-Cloning Site - Region in plasmid that has been engineered to contain recognition sites of a number of restriction nucleases (restriction enzymes)
-This means the plasmid was given specific nitrogen bases (in palindromic sequence, see last note, to allow enzymes to cut sequence at this point)
The Plasmid:
- Plasmids are small double stranded circular DNA molecules that exist within many bacteria.
- They range in size from 1000 - 200 000 base pairs.
- A plasmid can have foreign genes inserted into its structure which will then be read by the bacteria, and replicated.
- Plasmids are beneficial to the bacteria because they may contain genes that will help the bacteria to survive. For example, a resistance to heavy metals such as lead or mercury may be kept within a plasmid.
On the above plasmid there are 7 different recognition sites.
for a plasmid with only two recognition sites, the following choices can be made:
- Use one type of restriction enzyme to cut plasmid at one location(EcoRI for exapmple)
- Use a differnt restriction enzyme to cut plasmid at a different location (Hind III for example)
- Use both restriction enzymes to cut plasmid at both recognition sites simutaneously
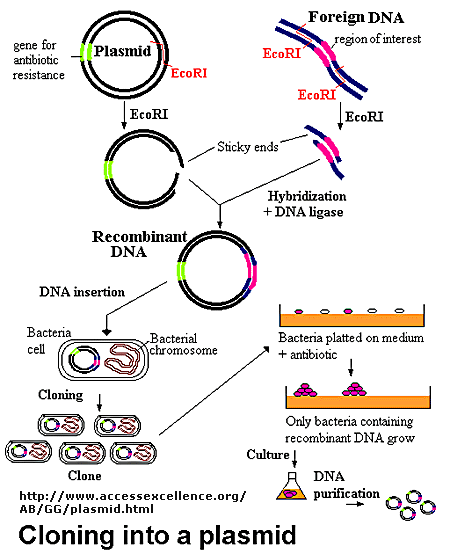
- When a plasmid is cut, a new gene is inserted into the solution containing the plasmid. The plasmid base pairs and the sticky ends of the inserted gene will anneal and link together creating a new plasmid with the gene located within this new plasmid.
No comments:
Post a Comment